One of the common problems with Sanger sequencing is samples with high salt concentration. In general, one could talk about inhibitors as such, but salts are probably the most common case. If you send us such samples, you will probably not be very satisfied with the quality of the result. In this article, we will take a closer look at why this is the case and, most importantly, what can be done about it.
Clients of our sequencing laboratory often require the preparation of sequencing libraries using rRNA depletion (ribodepletion), ie including rRNA removal. As we sometimes find that the results of rRNA removal are unsatisfactory, we performed tests described below.

Although state-of-the-art Next-Generation sequencing (NGS) technologies generate an adequate amount of sufficient quality data for de novo genome assembly of organisms for which the reference sequence is not known yet, the genome assembly process itself is still a major challenge for bioinformaticians. In this article, we will try to outline the issue of assembly and show possible solutions.

Sanger sequencing is associated with certain technical limitations, and one of the most important is read length. Typically, even with a perfectly flawless design, not more than a roughly 1,100 bases can be expected. But what is the lower limit of how long or rather short templates can be to be able to successfully sequence them? And can we sequence from the first base just after the sequencing primer? We will look at this issue in this article.

Illumina sequencing technology is commonly referred to as short-read technology. In practice this means that it typically produces reads of hundreds of bases in length. Its current technological maximum in terms of read length is a kit for the MiSeq system that allows reading up to 600 b (2x300 setting). In this article, we have taken a closer look at some of the problems associated with this type of sequencing.

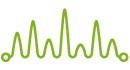
When sequencing PCR products, the fluorescent signal sometimes drops sharply and the PCR product sequence is not read completely. A typical cause of this problem is the presence of various inhibitors in templates of sequencing reactions.

In the last article we have described some features and consequent limitations of Sequel sequencer (Pacific Biosciences). If you are looking for an alternative to this long read technology, you might be considering the technology of Oxford Nanopore.

Pacific Biosciences SMRT sequencing technology enables sequencing of fragments of both tens of kilobase and hundreds of bases in length.

Illumina sequencing technology is currently dominating the market. It offers a number of advantages over other technologies but unfortunately this does not mean that it is completely free of imperfections.

Just as few people go to the theater or a sports match without knowing what is on the program, it is normal for any other service you intend to order to have at least some idea in advance of what you can expect from the service provider, especially in terms of QUALITY, SPEED and PRICE. This of course also applies to DNA sequencing. It is understandable that most clients would like all of these three criteria to be met at 100% at the same time, but we do say that this is not basically possible.

You may have noticed the recent announcement of ThermoFisher Scientific (Sep 2020) communicating the end-of-life of popular and widely used Applied Biosystems 3130/xl sequencers.

Our constant search for the best and most appropriate solutions to various experimental designs has led us to the introduction of the GemCode technology into our portfolio of Next-Generation sequencing services. In this post, we are happy to announce its availability to our clients and provide its basic description.

If I had a nickel for every time we answered the “How should I quantify my DNA or RNA for NGS library prep?” question, I wouldn’t need to be writing this article. I’d be on a boat in the Mediterranean. But since I’m not on a boat anywhere, I thought I could perhaps collect several good reasons why using Nanodrop is not really the best way of doing this.

For successful sequencing analysis it is crucial to use optimal template concentration (as described in our guidelines for sample preparation). If you do not have required amount of template, the total volume of a sample can be lowered but the template to primer ratio must be kept. The problem of low template concentration cannot definitely be solved by only increasing the total sample volume...

Routinely obtained sequence electropherograms typically start with unreliable (unreadable) data just at the beginning of a sequence. Although we should theoretically read the first base after the sequencing primer, there are often errors or truncations just behind the primer ...

Here and there we do receive a request to sequence large templates (i.e. chromosomal DNA, BACs, cosmids) directly. We can do this but similarly as for standard samples where short pcr products or plasmids serve as templates also here the key prerequisite is the template amount.

Results of Sanger sequencing are provided in three types of file formats and not every user of our services is familiar with efficient usage of these data files. Here are brief instructions how to use them properly.

In a relative quantification experiment, what you are most likely interested in is to compare the expression level of a particular gene among different samples. The most common way of correcting variations in the target nucleic acids input amount among samples is ...

Have you ever seen the Applied Biosystems 7900HT Real-Time PCR system? Without really knowing what you are looking at, you probably didn’t like it. In my opinion this is the ugliest qPCR instrument ever, grey, huge, heavy and noisy.

Many of our clients require a regular preventive maintenance. Belonging among them or not, have you ever thought about how you know the maintenance was performed properly?

One of our loyal customers reminded me of a letter he received recently from Life Technologies bringing up a topic dear to my heart – independent service providers! If you’ve been lucky enough to receive this letter too, I would like to take a moment to address concerns you may have.

Have you ever ordered sequencing analysis of a plasmid or PCR product and as a result received the DNA sequence that suddenly stops? One of the possible causes can be that in your template complex secondary structures are being formed, most often hairpins.

Are you wondering why you should send samples to us? Please read our 10 good reasons to answer this question!

Assuming we have good signals and read length is as expected which means we have successfully solved all issues mentioned in previous two parts of this post, we can still be far away from seeing nice data. This third part deals with the problem of having peaks overlapping other peaks. The sequence is not readable.

In the first part we discussed how to troubleshoot no or low signals. Basically empty electropherograms. Very frustrating. Now, I focus on another frequent result – you obtain some signals but the height of the DNA sequencing peaks diminished rapidly. A read length is very short.

It is not the aim of this post to provide a detailed description of all problems you may observe when evaluating your sequencing results. Instead, I focus on most frequent problems and recommend some steps to take for your consideration. Additionally, I am not covering issues related to instruments and sequencing reagents used in our (or any other) sequencing lab because first it is our responsibility to secure a problem-free sample processing on the instrument and second, from the user‘s point of view, it is of low interest because you cannot influence it anyway.

A very frequent scenario when cleaning up your samples (plasmids or PCR products) for DNA sequencing is to load your DNA to silica-based membrane in spin columns. Unfortunately some columns (even those of established manufacturers) may not have a really favorable shape and consequently your DNA gets dirty during purification.

The technology of next-generation sequencing produces huge amounts of data compared to Sanger technology. Its volume naturally depends on the design of the experiment, but primarily on the output capacity of the instrument. In principle, it is always necessary to deal with the transfer of large amounts of data into a form which enables their effective processing to allow a deeper analysis of the sequences obtained which is the very aim of the experiment.