Some people think they can't handle Next-Generation Sequencing data analysis. They are wrong!
From the broader perspective, NGS data analysis is a two-step process only. First is quality control to get the best data set available and simply said it is always the same. Second is of course the application-specific pipeline. This workshop will teach you to make the first step yourself. After all this is where you always begin. Its result will be a perfect mastery of NGS data quality control.
Join our in-person workshop and learn by doing!
You will get acquainted with various formats of NGS data, learn about specific advantages and disadvantages of sequencing technologies and understand the background of individual processes used to modify NGS data. After this three-day workshop, you will not become a bioinformatician, but you will be well on your way to becoming a qualified partner for bioinformaticians in your team. And with a great data set in your hand, by the way.
Day 1 - Know your environment
- General introduction to NGS data
- Overview of NGS data formats
- Introduction to the Linux system
- Navigation and basic commands in the Terminal
Day 2 - NGS data quality control
- Standard commands in the Terminal to process NGS data
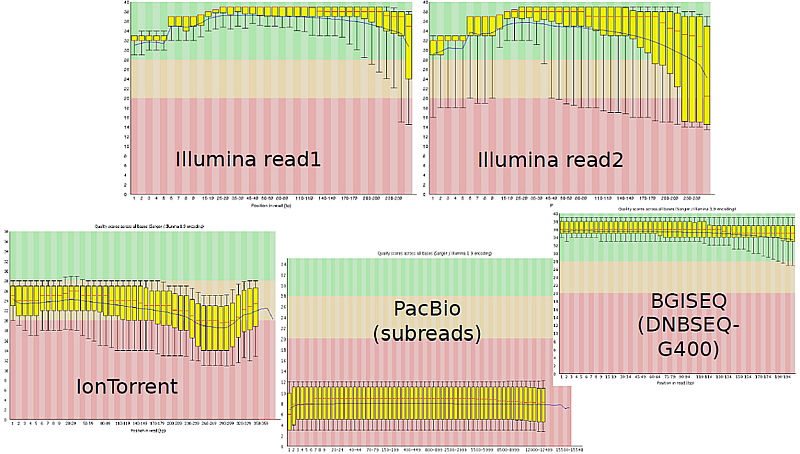
- Visualization of the quality of raw NGS data
- Pre-processing of short-reads FASTA/FASTQ files using various programs
Day 3 - Get the best data set
- Read mapping to the reference sequence
- Visualization and evaluation of mapping quality
- Identification and removal of duplicate reads
- Identification of unmapped reads
Who Should Attend?
- Biologists, bioinformaticians, healthcare professionals and laboratory workers interested in analyzing NGS data
- Great for beginners
- Suitable also if you want to deepen or revive your knowledge
What will you learn?
- Working in Ubuntu command line
- Evaluating quality of NGS data
- Basic operations with NGS data - removing reads with low quality, artifacts, duplication and contamination
- Viewing results
- In brief, you will learn to get high quality dataset for downstream application-specific pipeline.
Prerequisites
- This is a hands-on computer workshop with presentations and practical demonstrations.
- A necessary prerequisite is computer literacy and basic knowledge of molecular biology (DNA, RNA, gene expression, PCR). Knowledge of BASH and the terminal is not required, but is an advantage.
General information
- Language: English
- Printed presentations are provided for in-person events, pdfs for online events (will be emailed to you a few days before the online event commences).
- For in-person events lunches and refreshments are included.
- All tools we use during our events are open-source (free). We never use licenced software unless explicitely stated.
- All participants of our hands-on workshops perform important steps of data analysis themselves, each has their own computer we provide. Using your own computers and / or data files during our data analysis workshops is not possible.
- Time is always CET.
- Lodging, travel, and other incidental expenses (if applicable) are the responsibility of attendees.
- No laboratory experiments are conducted during our events.
- For all our events you will receive a certificate of completion.
About us
This event is organised by SEQme company. We are an NGS-service provider with laboratories located in the Czech Republic and serving several hundreds of clients every year.
We organize courses and workshops since 2012. Our team of speakers is comprised of our employees, NGS lab specialists, data analysts and bioinformaticians. For selected events from our portfolio we work together also with our external partners and collaborators, usually bioinformaticians with deep knowledge of particular application-specific pipelines.

Head of our Courses & Workshops programme: Stepan Stoces
Please send your inquiries to ngs@seqme.eu
We can make it a turnkey event for your team only. Do not hesitate to contact us!