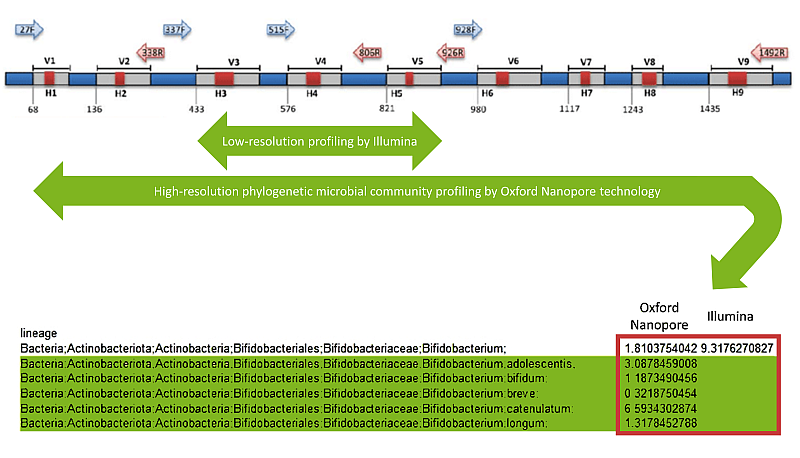
Is sequencing hypervariable regions by Illumina good enough for you? And do you know what you are losing?
This workshop is your gateway to mastering Oxford Nanopore (ONT) metagenomic sequencing, a cutting-edge technology that allows for long reads sequencing, providing unparalleled insights into the genomic makeup of complex microbial ecosystems. Get ready for an immersive dive into the world of metagenomic analysis with this full-length metagenomic proficiency workshop!
This workshop has been designed to equip participants with the skills needed to navigate the challenges of metagenomic studies using long reads.
Day 1 - Introduction to ONT and Metagenomics
- Overview of Oxford Nanopore sequencing technology
- Metagenomic Analysis Basics
- Introduction to basic operations in linux
- Fundamentals of metagenomic analysis
- Types of metagenomic studies and their objectives
- Advantages and applications in metagenomics
- Practical aspects of ONT sequencing
Day 2: Advanced Metagenomic Analysis
- ONT Workflow Optimization - Tips and tricks for optimizing ONT workflows in metagenomic studies
- Advanced Data Analysis Techniques
- Exploration of metagenomic data analysis tools and techniques
- Analyze real-world metagenomic datasets
Day 3: Hands-On Metagenomic Data Analysis
- ONT amplicon-based metagenomic analysis
- Case studies
- ONT whole-genome metagenomic analysis
- Case studies
Who Should Attend?
- Biologists, bioinformaticians, healthcare professionals and laboratory workers engaged in metagenomic analysis or those seeking to enhance their proficiency with ONT technology.
- Whether you are a novice or experienced researcher, this workshop caters to all levels.
- Suitable also if you want to deepen or revive your knowledge
What will you learn?
- ONT Proficiency: You will gain expertise in analyzing ONT data
- Metagenomic Analysis Techniques: You will learn essential techniques for analyzing metagenomic data
- Interpretation Skills: You will develop the ability to interpret and draw meaningful conclusions from metagenomic datasets
- Best Practices: You will understand the best practices for experimental design and data analysis in metagenomics
Prerequisites
- This is a hands-on computer workshop with presentations and practical demonstrations.
- Basic knowledge of working in terminal is recommended. Consider attending our NGS Data Analysis for Beginners Workshop.
General information
- Language: English
- Printed presentations are provided for in-person events, pdfs for online events (will be emailed to you a few days before the online event commences).
- For in-person events lunches and refreshments are included.
- All tools we use during our events are open-source (free). We never use licenced software unless explicitely stated.
- All participants of our hands-on workshops perform important steps of data analysis themselves, each has their own computer we provide. Using your own computers and / or data files during our data analysis workshops is not possible.
- Time is always CET.
- Lodging, travel, and other incidental expenses (if applicable) are the responsibility of attendees.
- No laboratory experiments are conducted during our events.
- For all our events you will receive a certificate of completion.
About us
This event is organised by SEQme company. We are an NGS-service provider with laboratories located in the Czech Republic and serving several hundreds of clients every year.
We organize courses and workshops since 2012. Our team of speakers is comprised of our employees, NGS lab specialists, data analysts and bioinformaticians. For selected events from our portfolio we work together also with our external partners and collaborators, usually bioinformaticians with deep knowledge of particular application-specific pipelines.

Head of our Courses & Workshops programme: Stepan Stoces
Please send your inquiries to ngs@seqme.eu
We can make it a turnkey event for your team only. Do not hesitate to contact us!