
Cost effective sequencing library preparation strategy for amplicons and other DNA fragments of your interest
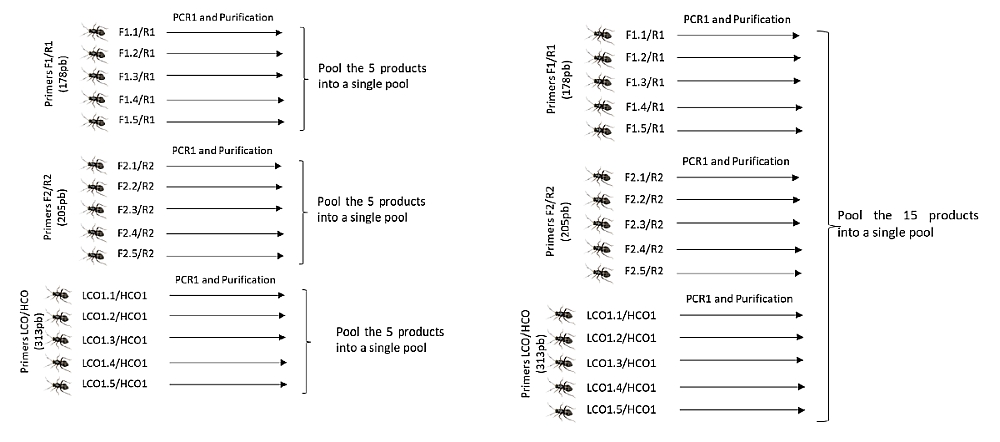
Three samples (pools) containing 5 amplicons each (left) or a more sophisticated design where 15 amplicons are pooled into one sample (right). If the library contains, for example, 94 samples, 470 (left) or 1410 (right) amplicons can be sequenced in this way. The data is demultiplexed into individual samples (94 files) upon delivery. Further differentiation takes place according to the length of the product, the sequence of primers or other identifiers used. Image courtesy of Mr. Luis Perera Fernández.
|
Should you need a really low-cost sequencing strategy for hundreds or even thousands of DNA fragments at a time, this Tailed Amplicon approach is the right choice for you. Although its name mentions amplicons, it is suitable for dsDNA fragments bearing specific tails at both 5´ and 3´ends, obtained not necessarily by PCR exclusively. In fact, it is compatible with DNA fragments generated by tagmentase cleavage (Illumina Tagment DNA TDE1 Enzyme) or DNA fragments having specific tails ligated. In any case, DNA fragments supplied must bear specific tails as outlined below! Why? Because in our sequencing lab, these specific tails are utilized in order to add another set of tails (adapters) needed for sequencing. At this step, unique index combination chosen from our large stock is introduced and allows high level of multiplexing. This approach enables you to focus on any region of your interest. Also it leaves the initial sample preparation under your full control enabling you to work with reagents of your choice but without the need of purchasing large number of indexed adaptors. Instead, we supply these and prepare the sequencing library for subsequent sequencing using any Illumina platform we offer. |
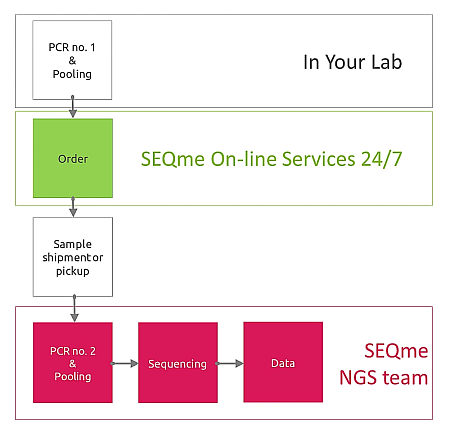 |
Step 1 in YOUR lab - You perform PCR to amplify regions of interest and pool all amplicons from the same sample into a single pool. For example, should you have 94 samples and 5 different amplicons per sample, you perform 5 x 94 PCR reactions and pool amplicons per sample. Samples must be further purified and diluted to a uniform concentration according to the valid Sample submission guidelines.
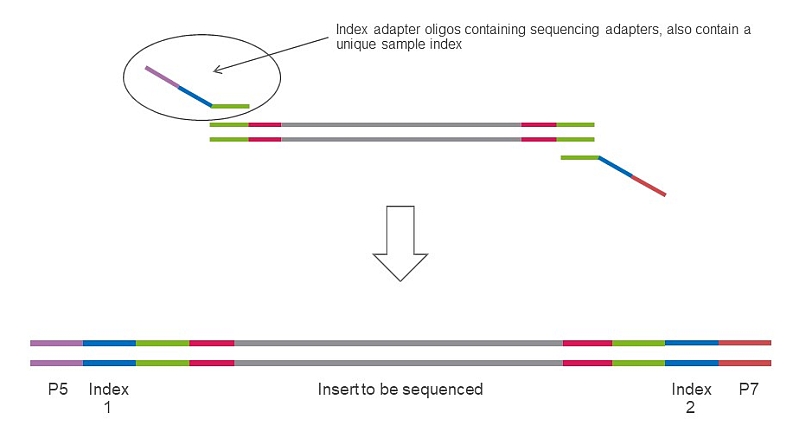
Step 2 in SEQme NGS lab - We perform 2nd PCR step to add sequencing indices and adapters, perform all necessary QC steps and pool all 94 samples into a single library and proceed with sequencing.
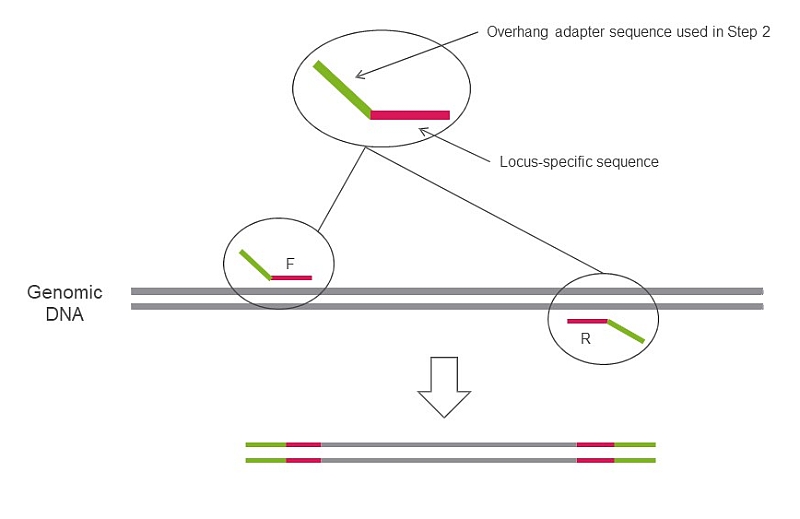
Append to 5’ end of your primers:
You perform 1st PCR step using primers above. Then, in our lab your samples will be processed as follows:
Approx. 4 weeks (data analysis not included)
Follow our Sample submission guidelines.
The analysis can be ordered and commissioned online as a package for 24, 48, 94 or 190 samples. If you want to order an analysis of a different number of samples (eg only a few samples or, conversely, thousands), contact us to prepare an individual calculation.