
Rapid and cost-effective solution for exome analysis based on the most advanced design and most proven SureSelect technology
Whole exome sequencing (WES) is the most effective way of studying coding regions of the genome. For example human exome consists of about 20.000 protein-coding genes, constituting only about 1,5% of the genome but up to 85% of disease-causing mutations can be located within exome. Exome sequencing enables to identify common variants (SNV), copy-number variants (CNV), small insertions or deletions (InDel) and even rare de-novo mutations.
In our sequencing lab we utilize probe hybridization technology developed by Agilent Technologies, Inc. that has proven to be very efficient and reliable.
As a standard, we offer whole-genome sequencing of human and mouse DNA samples; for other organisms contact us if you are interested.
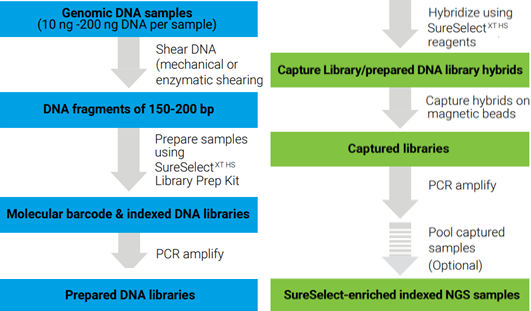
SureSelect XT HS enrichment system (Agilent Technologies) utilizies RNA probes to hybridize to target DNA regions. The main advantages of this technology are:
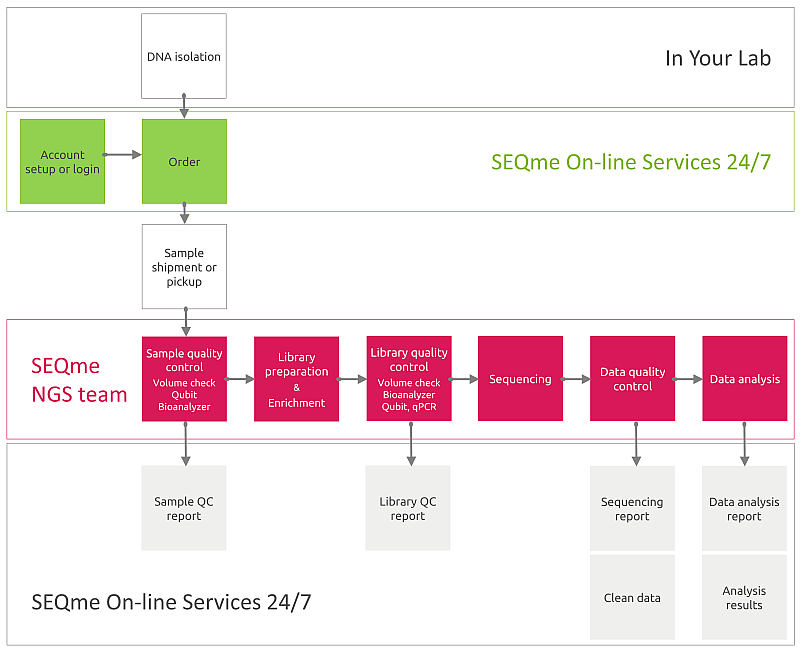
You perform DNA isolation. Then, in our lab your samples will be processed as follows:
The sequences obtained will be sorted according to the combination of indexes into files representing individual samples and analysis of sequencing quality indicators such as number and length of sequences, phred score, %GC, duplication level, etc. will be performed.
As output you will receive data in FASTQ format divided into files according to individual samples. Upon request, the outputs can be processed in another form. Please specify this when ordering.
We can perform also data analysis including data QC, mapping to the reference sequence, variant calling, filtering of variants and annotation. You will receive a comprehensive data analysis report.
Approx. 4 weeks (data analysis not included)
Follow our Sample submission guidelines.