In the last article we have described some features and consequent limitations of Sequel sequencer (Pacific Biosciences). If you are looking for an alternative to this long read technology, you might be considering the technology of Oxford Nanopore, which currently offers several sequencers, essentially different in capacity. The smallest device is the MinION system. SEQme offers sequencing on GridION, the larger brother of the popular MinION.
Oxford Nanopore technology is developing really fast, and its advancements leave no doubt that it has advanced beyond the "untrustworthy" stage of generating low-quality data. We strongly recommend you to consider it, of course, especially in cases where you are no longer satisfied with short reads generated by the market leader Illumina. However, there are still some limitations at Oxford Nanopore and the following article tries to summarize the main ones.
As with the Sequel system, the sequencing output (data volume) is significantly determined by the length of the input fragments. Amplicons, transcripts or genomic DNA of different lengths can be sequenced. In addition, sequencing on the GridION system is very sensitive to specific inhibitors, which can kill smaller or larger number of nanopores and thus reduce sequencing output.
Under optimal conditions, the output capacity of Oxford Nanopore systems is determined by the number of active pores and sequencing time. Sequencing time can be divided among multiple samples within the maximum sequencing time. In practice, this means that the sequencing can be started and when we think we have enough data, it can be stopped, the sequencing chip (flowcell) washed, and we can proceed with the sequencing of the next sample on the same chip up to the total maximum time it can handle. After some time, sequencing loses efficiency and is no longer worth continuing.
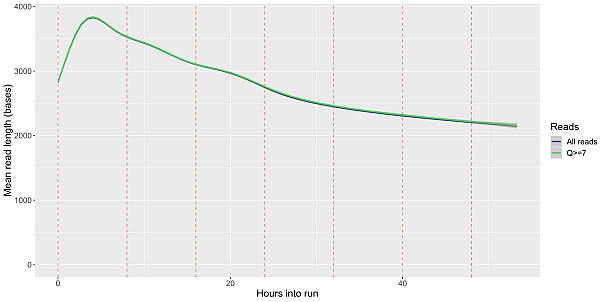
Decrease of sequencing output over time for MinION system
The sequencing time, i.e. the time for which the pores are active, is subject to a very progressive development, which translates into the growth of the maximum sequencing output. Together with another rapidly evolving parameter, which is increasing sequencing speed, there is a continuous increase in the capacity of sequencing chips:
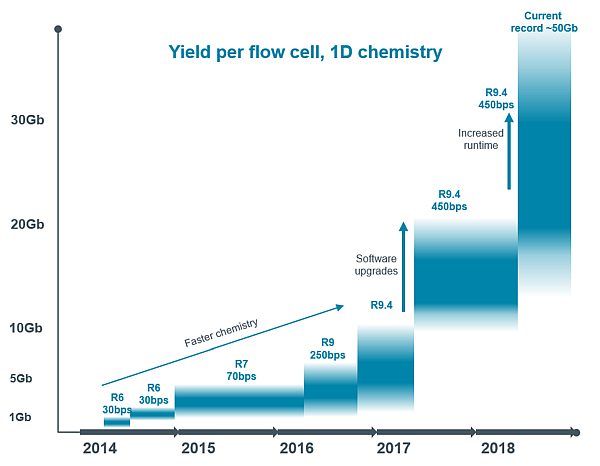
Minion / GridION chip capacity development
Due to this very rapid development, I do not list specific figures here. However, irrespective of the speed of development, the variables that affect sequencing output are still the same and include in particular:
Sequencing accuracy of this technology has rightly a bad reputation since its onset, but in this area too, Oxford Nanopore is pushing for continuous improvement. It went through several dead ends, which were 2D and 1D2 readings, which were aimed at obtaining a sequence from both complementary strands and comparing them to eliminate errors. In both cases, however, the technology faced the problem that only a low percentage of reads was the product of these strategies, while most reads were so-called 1Ds. Both strategies were abandoned, although they helped to improve reading accuracy.
A strategy that has proven successful at Oxford Nanopore, and which leads to a continuous improvement in reading quality, is the development of algorithms that convert the signal to basis = basecalling. Unique to other sequencing technologies is the possibility of storing data in the form of signal recording (although this data is very large) and repeating basecalling with the latest software version. So you can actually read old data, much like reading a book in your youth that you did not understand and which you will return to later.
All these features and the rapid technological development lead us to recommend GridION and technology as such with no hesitation. It is true that the accuracy is just over 90%, but still it is a technology that is already fully usable for many applications, especially in combination with data produced by another technology (typically Illumina).
Note: Images by Oxford Nanopore, modified.
NGS Lab, ngs@seqme.eu