Our constant search for the best and most appropriate solutions to various experimental designs has led us to the introduction of the GemCode technology into our portfolio of Next-Generation sequencing services. In this post, we are happy to announce its availability to our clients and provide its basic description.
The GemCode technology has been developed by the 10xGenomics company, based in Pleasanton, California and is being represented by an innovative system that transforms short-read sequencing technologies - Chromium System.
The key highlights of the Chromium system are:
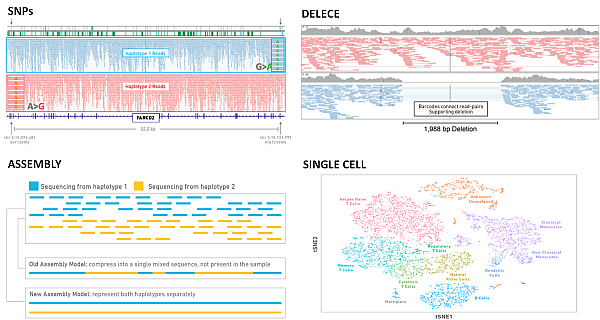
In the picture below you can see some application examples (Zheng et al. 2017):
Technology
The GEM technology (Gel bead in Emulsion) enables to capture long DNA molecules or RNA from single cells into droplets where they are turned into droplet-specific (or source molecule/cell-specific) barcoded short fragments. These fragments when sequenced are used for reconstruction of its source molecule (DNA) or for analyzing transcriptome of each single cell (RNA).
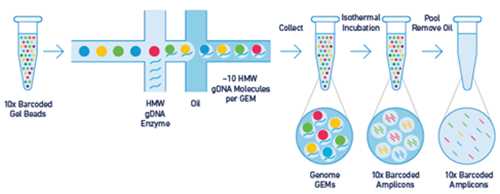
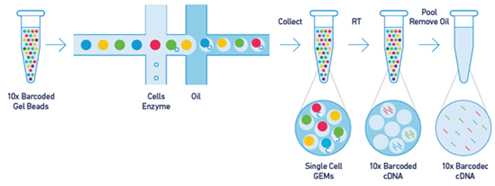
Long molecules capture (left) and Cells capture (right)
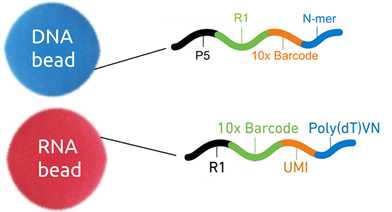
In droplets, long molecules (DNA or RNA) are amplified using primer extension. These molecules are not extended to its full length but only to the length compatible with Illumina sequencers. Primers used for extension, at first attached to the surface of a gel bead, contain the annealing part, 14 bases long barcode and Illumina read one primer specific sequence. In case of DNA, primers also include a part of P5 adaptor and in case of RNA, the UMI part (Unique Molecular Identifier) which might be used to reduce the quantitative bias introduced by replication.
Instrumentation
Library construction using GEM is performed using Chromium controller instrument which might be best described as a hybrid between a flow cytometer and an emulsion PCR machine.

The Chromium controller is using 750,000 different barcodes. Each droplet contains several molecules and thus it gives the capacity of reconstructing 5 – 10 million of 50 kb molecules or to analyze several hundred thousands of individual cells. It is very important to point out that the GemCode technology leverages existing short-read sequencers (Illumina, see below) and is easily integrated into existing laboratory workflows.
Data analysis
For reconstruction of source molecules The Long Ranger open-source software is being used. Other software packages for downstream analysis such as Supernova for de-novo assembly, Cell Ranger for transcriptomics or Loupe for visualization are also freely available. Genome analytical pipelines also use specially developed aligner 10xLariat which is tuned to data generated using this technology.
Technical specifications:
If you are interested in using this technology in your research, please do not hesitate to contact us.
NGS Lab, ngs@seqme.eu
Literature:
Zheng GX, Terry JM, Belgrader P, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049.